With the discharge of platforms like DALL-E 2 and Midjourney, diffusion generative fashions have achieved mainstream reputation, owing to their capability to generate a collection of absurd, breathtaking, and sometimes meme-worthy photos from textual content prompts like “teddy bears engaged on new AI analysis on the moon within the Nineteen Eighties.” However a staff of researchers at MIT’s Abdul Latif Jameel Clinic for Machine Studying in Well being (Jameel Clinic) thinks there may very well be extra to diffusion generative fashions than simply creating surreal photos — they might speed up the event of latest medication and scale back the probability of opposed unintended effects.
A paper introducing this new molecular docking mannequin, referred to as DiffDock, will likely be introduced on the eleventh Worldwide Convention on Studying Representations. The mannequin’s distinctive method to computational drug design is a paradigm shift from present state-of-the-art instruments that the majority pharmaceutical firms use, presenting a significant alternative for an overhaul of the normal drug growth pipeline.
Medicine sometimes perform by interacting with the proteins that make up our our bodies, or proteins of micro organism and viruses. Molecular docking was developed to realize perception into these interactions by predicting the atomic 3D coordinates with which a ligand (i.e., drug molecule) and protein may bind collectively.
Whereas molecular docking has led to the profitable identification of medicine that now deal with HIV and most cancers, with every drug averaging a decade of growth time and 90 p.c of drug candidates failing expensive medical trials (most research estimate common drug growth prices to be round $1 billion to over $2 billion per drug), it’s no surprise that researchers are on the lookout for sooner, extra environment friendly methods to sift by potential drug molecules.
Presently, most molecular docking instruments used for in-silico drug design take a “sampling and scoring” method, trying to find a ligand “pose” that most closely fits the protein pocket. This time-consuming course of evaluates a lot of totally different poses, then scores them based mostly on how effectively the ligand binds to the protein.
In earlier deep-learning options, molecular docking is handled as a regression drawback. In different phrases, “it assumes that you’ve a single goal that you just’re attempting to optimize for and there’s a single proper reply,” says Gabriele Corso, co-author and second-year MIT PhD pupil in electrical engineering and laptop science who’s an affiliate of the MIT Laptop Sciences and Synthetic Intelligence Laboratory (CSAIL). “With generative modeling, you assume that there’s a distribution of doable solutions — that is vital within the presence of uncertainty.”
“As an alternative of a single prediction as beforehand, you now enable a number of poses to be predicted, and every one with a distinct chance,” provides Hannes Stärk, co-author and first-year MIT PhD pupil in electrical engineering and laptop science who’s an affiliate of the MIT Laptop Sciences and Synthetic Intelligence Laboratory (CSAIL). Because of this, the mannequin would not must compromise in trying to reach at a single conclusion, which is usually a recipe for failure.
To grasp how diffusion generative fashions work, it’s useful to elucidate them based mostly on image-generating diffusion fashions. Right here, diffusion fashions steadily add random noise to a 2D picture by a collection of steps, destroying the info within the picture till it turns into nothing however grainy static. A neural community is then skilled to get well the unique picture by reversing this noising course of. The mannequin can then generate new knowledge by ranging from a random configuration and iteratively eradicating the noise.
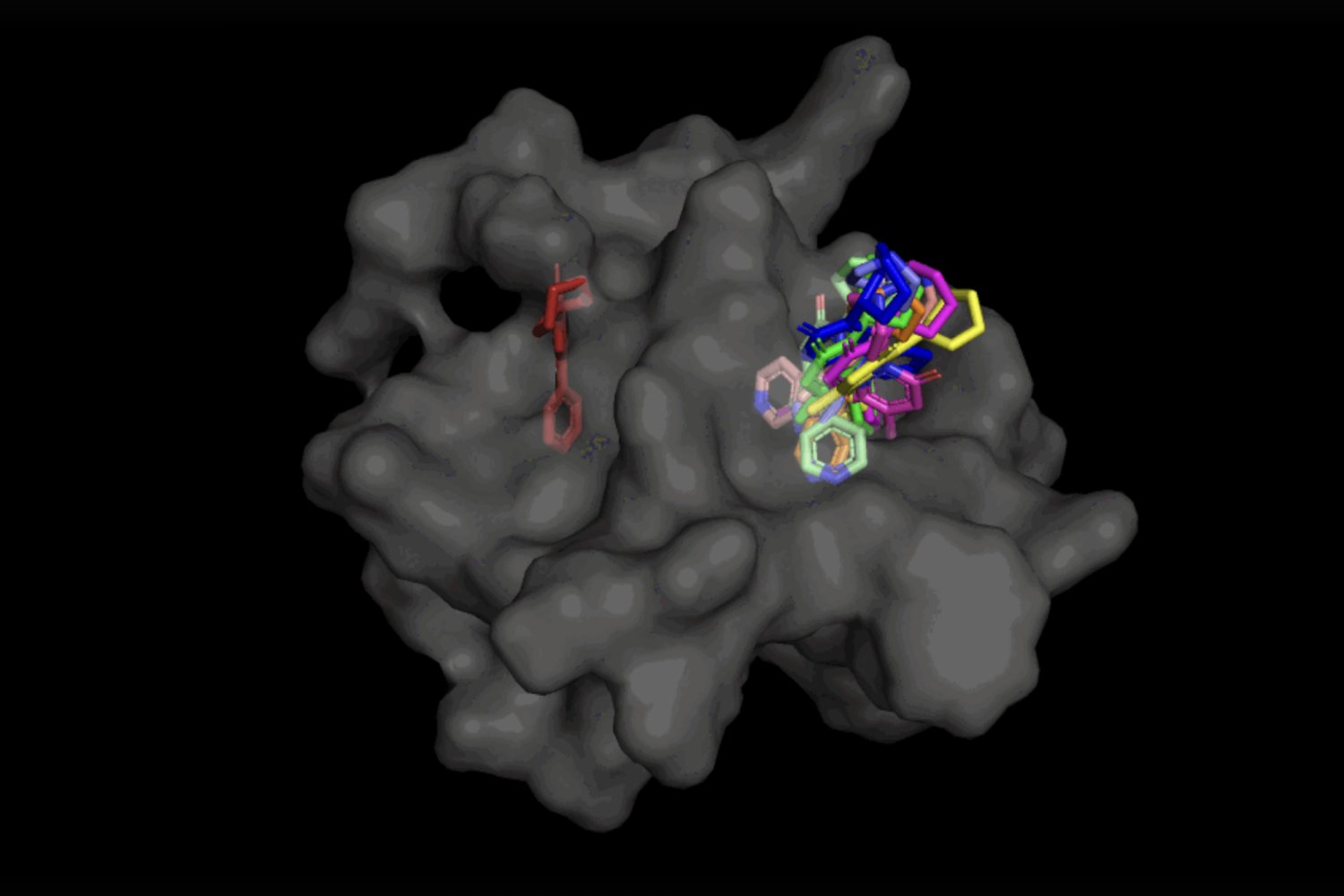
Within the case of DiffDock, after being skilled on quite a lot of ligand and protein poses, the mannequin is ready to efficiently determine a number of binding websites on proteins that it has by no means encountered earlier than. As an alternative of producing new picture knowledge, it generates new 3D coordinates that assist the ligand discover potential angles that will enable it to suit into the protein pocket.
This “blind docking” method creates new alternatives to reap the benefits of AlphaFold 2 (2020), DeepMind’s well-known protein folding AI mannequin. Since AlphaFold 1’s preliminary launch in 2018, there was a substantial amount of pleasure within the analysis group over the potential of AlphaFold’s computationally folded protein constructions to assist determine new drug mechanisms of motion. However state-of-the-art molecular docking instruments have but to reveal that their efficiency in binding ligands to computationally predicted constructions is any higher than random likelihood.
Not solely is DiffDock considerably extra correct than earlier approaches to conventional docking benchmarks, because of its capability to purpose at the next scale and implicitly mannequin a few of the protein flexibility, DiffDock maintains excessive efficiency, whilst different docking fashions start to fail. Within the extra sensible situation involving the usage of computationally generated unbound protein constructions, DiffDock locations 22 p.c of its predictions inside 2 angstroms (extensively thought-about to be the brink for an correct pose, 1Å corresponds to at least one over 10 billion meters), greater than double different docking fashions barely hovering over 10 p.c for some and dropping as little as 1.7 p.c.
These enhancements create a brand new panorama of alternatives for organic analysis and drug discovery. As an illustration, many medication are discovered through a course of generally known as phenotypic screening, by which researchers observe the results of a given drug on a illness with out understanding which proteins the drug is appearing upon. Discovering the mechanism of motion of the drug is then vital to understanding how the drug could be improved and its potential unintended effects. This course of, generally known as “reverse screening,” could be extraordinarily difficult and dear, however a mixture of protein folding strategies and DiffDock might enable performing a big a part of the method in silico, permitting potential “off-target” unintended effects to be recognized early on earlier than medical trials happen.
“DiffDock makes drug goal identification rather more doable. Earlier than, one needed to do laborious and dear experiments (months to years) with every protein to outline the drug docking. However now, one can display screen many proteins and do the triaging just about in a day,” Tim Peterson, an assistant professor on the College of Washington St. Louis Faculty of Drugs, says. Peterson used DiffDock to characterize the mechanism of motion of a novel drug candidate treating aging-related ailments in a latest paper. “There’s a very ‘destiny loves irony’ side that Eroom’s regulation — that drug discovery takes longer and prices more cash every year — is being solved by its namesake Moore’s regulation — that computer systems get sooner and cheaper every year — utilizing instruments resembling DiffDock.”
This work was carried out by MIT PhD college students Gabriele Corso, Hannes Stärk, and Bowen Jing, and their advisors, Professor Regina Barzilay and Professor Tommi Jaakkola, and was supported by the Machine Studying for Pharmaceutical Discovery and Synthesis consortium, the Jameel Clinic, the DTRA Discovery of Medical Countermeasures Towards New and Rising Threats program, the DARPA Accelerated Molecular Discovery program, the Sanofi Computational Antibody Design grant, and a Division of Power Computational Science Graduate Fellowship.